Chapter 7 A Deeper Dive into Viral Genomic Complexity
Gytis Dudas
Vilnius University, Vilnius, Lithuania
Thus far in this book, we’ve discussed the principles of genomic epidemiology, and how genomic diversity accrues under the assumption of clonal evolution. This is both a matter of the pathogens we’ve focused on up to this point – viruses – and also a matter of the techniques and analysis we’ve discussed – mutations accruing during the process of error-prone replication, using the rate of mutation accrual as a molecular clock, and visualizing those patterns of shared and unique mutations with phylogenetic trees. While looking at the accrual of genomic diversity through clonal evolution is fundamental to the topics and techniques of genomic epidemiology, and useful across assorted pathogens, it is not the only way in which pathogens generate genomic diversity. In this chapter, we discuss additional mechanisms by which viruses can accrue genomic diversity, and different viral genome organizations, with a particular focus on how that affects genomic epidemiological analysis and inference.
7.1 Recombination
Typically, when we talk about evolution, we consider the process by which an organism’s genetic material is copied with random errors (mutations) and passed on to their progeny. Those progeny then have unequal chances of surviving and passing on that inherited variation, with additional mutations sprinkled in, to their offspring. Most probabilistic phylogenetic methods (like ML or Bayesian approaches) aim to recapitulate this process by inferring whether sequences are more likely to share a mutation because the same mutation happened in other sequences, or because two or more sequences inherited the mutation from a common ancestor. The straightforward model of evolution where genetic material is inherited, mutated during replication, and then passed onto progeny is called clonal evolution, and it is how the vast majority of life evolves. However, recombination is another way to generate genetic variation, and it is a frequent addition on top of the process of clonal evolution.
To use an analogy, human language is often “inherited” wholesale by offspring during childhood largely unchanged, which resembles clonal evolution. Occasionally, if a child has contact with other languages, they may borrow words from that other language and intersperse it in their speech; that borrowing is similar to how recombination works. In the strictest sense, recombination happens when a strand of genetic material is broken and joined onto another. In complex organisms like animals, recombination is nearly universal and obligatory – during the production of gametes (meiosis), each chromosome we inherited whole from our parents is recombined, that is, a new chromosome is created by randomly “sampling” regions from one or the other parental chromosome. In non-eukaryotic microorganisms, recombination can integrate extraneous genetic material unrelated to anything in the host’s genome in what is known as horizontal gene transfer, which is a type of non-homologous recombination (non-homologous because the host and incoming genetic material are not related). We’ll talk more about this in Chapter 8. In RNA viruses specifically, nearly all recombination occurs between related sequences due to the template switching mechanism, whereby the RNA-dependent RNA polymerase dissociates from the original template it had been copying, together with the nascent RNA copy, and binds to a new template that is sufficiently complementary to the nascent copy to pair up and allow the continuation of RNA synthesis. The resulting RNA copy thus ends up being part original template and part new template.
Template switching likely takes place during all RNA virus infections, with the exception of negative sense single-stranded RNA viruses which effectively do not recombine. But in most cases, template switching will occur in a genetically homogenous infection in which the old template and the new template will be essentially the same genotype. Thus even with the process of template switching occurring, the resulting recombined sequences will not be distinguishable as such. This means that if you detect recombination in RNA viruses, likely the host had a genetically diverse infection, such as being co-infected with multiple different strains of a virus. Tools for detecting recombination are readily available but are not always well-explained or easy to interpret because they provide single numerical values like p-values, statistics, etc. Tests like these have a place in confirming recombination, but should not replace manual investigations of your sequence data for tell-tale signs of recombination.
What are these tell-tale signs? Firstly, you will likely observe an excessive number of repeat mutations; we refer to these as homoplasies. Walking through a toy example, imagine that at some point a virus has an A to G substitution at site 201 of its genome (A201G). If evolution proceeds only clonally, then we would expect that A201G mutation to occur and be passed on to the offspring. Notably, those offspring are descended from the same ancestor, and so they’ll share other parts of the genome as well. This means that the progeny that inherit the A201G mutation will likely cluster together closely in a phylogenetic tree, and a single instance of the A201G mutation that is then inherited by the group of related viruses will explain the phylogenetic pattern. However, if recombination is occurring, then a genetically diverse set of viruses could “pick up” the genetic material carrying the A201G mutation. On a tree, this would look as though many diverged groups of viruses all have that A201G mutation, even though the rest of their genomes are quite different. If the model strictly assumes clonal evolution, then the model’s only explanation for this A201G pattern across all of these different groups of viruses is to infer that the A201G mutation happened many times independently. And thus, we have excessive repeat instances of A201G: a homoplasy. For another example of homoplasies, we can return to our language analogy from earlier. Many languages, even non-Romance languages, have Latin words. If we assumed that language only evolved clonally, then we would look at these Latin words across all of the different languages, and we would say that they had been invented multiple times independently (this would be the homoplasy). Of course, the truth is that Latin words were not invented over and over again, but rather invented once in the Romance language family, and transferred by adoption to non-Romance languages.
Another feature of recombination is the spatial clustering of homoplasies along the sequence. This clustering occurs because recombination typically transfers stretches of genetic material, not just a single site. If the donor material is sufficiently diverged from its homologous counterpart in the recipient, then that stretch of genetic material will contain numerous mutations.
When unaccounted for, sufficiently recombined sequences used in conjunction with strictly clonal models of evolution lead to nonsensical inferences that can mislead the researcher or molecular epidemiologist. Judging how much recombination is too much, and when inferences are sabotaged beyond utility, is not easy and varies from situation to situation. Knowing the biology and ecology of the disease helps. If the pathogen is genetically homogenous, as is the case early in pandemics for example, or if co-infections are known to be rare, it can be argued that recombination is unlikely without even looking at the data. Other clues can also distinguish situations that produce similar signals to recombination from actual recombination. Recall that analysing recombinant sequences in phylogenetic models that are strictly clonal results in algorithms inferring that mutations transferred via recombination are recurring independently (homoplasies). While the recurrence of any given mutation (particularly in larger genomes) seems unlikely, the reality is that so many viral and bacterial progeny are produced during an infection, almost every possible mutation will occur. Thus in the presence of strong selective pressures to solve similar problems, recurrent changes are entirely expected. One of the best examples of this is E627K replacement in the PB2 protein of avian influenza A viruses jumping into humans. Virtually all human cases have this particular mutation, and with sufficient sampling of the virus in birds, it can be shown that this mutation occurs de novo in every single new jump into humans. Only recently was it shown that a specific interaction of the PB2 protein with ANP32A, a host protein that is sufficiently different between birds and humans, is at the root of this extremely strong selective pressure. In contrast, it is uncommon for synonymous mutations to be under similar selective pressures, and thus synonymous mutations are unlikely to recur and are a more believable marker of recombination.
One of the most thorough, unambiguous, and elegant demonstrations of recombination was performed by Jackson and colleagues23 during the SARS-CoV-2 pandemic. Due to a comprehensive and long-running genomic surveillance programme in the UK, it had been possible to identify SARS-CoV-2 genomes assigned to lineage B.1.1.7 (Alpha in WHO nomenclature) but which didn’t have all the mutations/Single Nucleotide Polymorphisms (SNPs) that B.1.1.7 should have. A closer examination of such genomes revealed that regions missing B.1.1.7 mutations had mutations typical of other lineages, strongly suggestive of recombination. Furthermore, at least four distinct types of recombinant genomes were found, each with multiple sampled genomes sequenced by different groups. This is an extremely useful finding since it suggests that this signal of recombination did not originate from sample contamination or sequencing error. Another important aspect demonstrated by the researchers in this study was that putative parental lineages co-circulated at the same time in similar geographic locations.
One quick test for recombination is to look at multiple alignment columns that only contain two alleles within the column, but have four possible haplotypes between all the columns. For example, in Figure 7.1, let’s take two columns of the alignment: the column at site 21255 (which contains either a C or a G), and the column at site 23063 (which contains either an A or T). These two columns span the recombination breakpoint. If we look at these two sites in the reference genome (grey), we see that the reference has 21255G and 23063A. The P1 Wales genome (B.1.177) has 21255C and 23063A. The P2 England genome (B.1.1.7) has 21255G and 23063T. The recombinant sequences (all Q Wales) have 21255C and 23063T. Thus, across the sequences we have all four possible haplotypes: GA, CA, GT, and CT. This pattern should cue us to the presence of recombination.
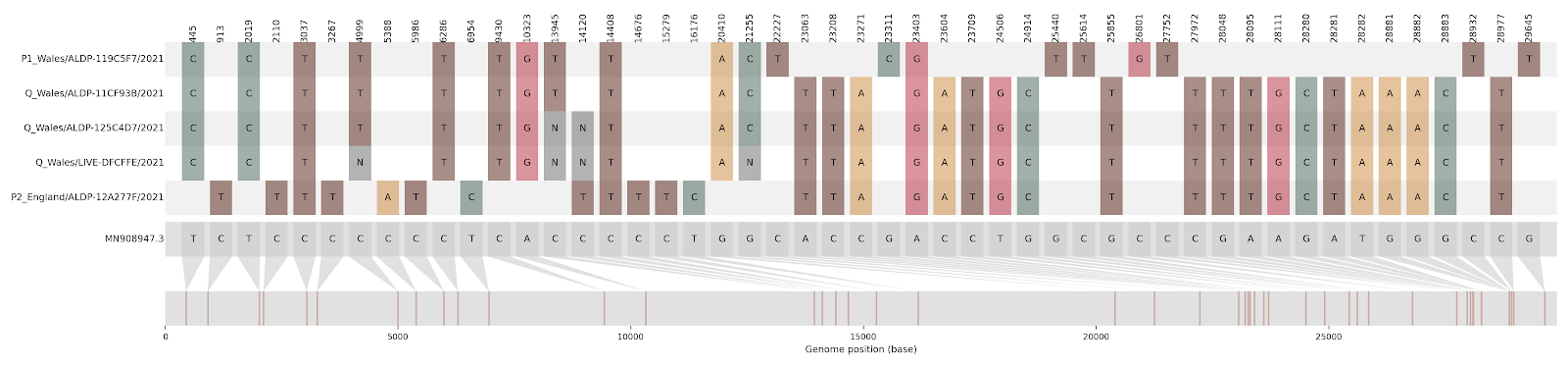
Figure 7.1: This condensed alignment shows mutations in the SARS-CoV-2 genome compared to the Wuhan-Hu-1 reference strain (shown in grey at the bottom, with mutation positions mapped below). The first and fifth sequences in the alignment (P1 Wales, and P2 England) are the non-recombinant “parent” sequences, while sequences two, three, and four (all Q Wales) represent the putative recombinant sequences. The P1 Wales virus is assigned Pango lineage B.1.177 and the P2 England virus is B.1.1.7. Looking at the alignment, you can see that the Q Wales sequences have SNPs characteristic of the B.1.177 virus in the first portion of the alignment, and then SNPs characteristic of B.1.1.7 in the latter portion of the alignment. The shift in which parent the Q Wales sequences look like, aka the recombination breakpoint, occurs between site 21255 and site 22227.
The logic of this simple test is that, when evolution only proceeds clonally, then one mutation arises first and the second mutation can only arise upon a genetic backdrop with the first mutation. This means that, if evolution is only occurring clonally, you shouldn’t see an instance where the “second” mutation occurs in the absence of the “first”.
7.2 Segmented Genomes and Reassortment
Several prominent groups of RNA viruses possess genomes that are segmented, meaning that their genomes are distributed across more than one piece of RNA. Groups with segmented genomes include orthomyxoviruses (influenza viruses with seven to eight segments being the best known), arena- and bunyaviruses (two to three segments, famous members include pathogens Lassa fever virus and Hantavirus), reoviruses (up to 11 segments, includes rota- and bluetongue viruses) and many others. Having a segmented genome can complicate many analyses in two ways.
Firstly, if you are performing metagenomic sequencing to characterise a novel segmented virus, it is usually easier to identify segments that are more conserved by homology, such as RNA-dependent RNA polymerases. It is usually much harder to reconstruct smaller and less evolutionarily constrained segments that do not resemble other sequences available on public databases. This problem may be circumvented if there are multiple independent samples of the same virus where the consistent co-occurrence of closely related stretches of sequence may be used to identify likely viral segments. Once these portions of the genome are identified as potential segments, it is not uncommon to carry out rapid amplification of cDNA ends (RACE) on putative segments to strengthen the hypothesis that they might be real segments. Many segmented viral groups have similar and partially complementary untranslated regions at the ends of each segment that help with genome packaging, and the presence and similarity of these may indicate their identity as segments belonging to the same genome.
The second complication associated with segmented genomes is reassortment. Reassortments can happen when two related but distinct segmented viruses co-infect the same cell. In the absence of incompatible genome packaging signals, new virions may package some combination of segments derived from both parental viruses. This often means that each segment evolves partially independently from its companion segments and the phylogenetic trees may differ markedly from segment to segment in a given collection of genomes. In some genomes (such as reoviruses), recombination may occur on top of reassortment within segments, complicating phylogenetic analysis even further. Fortunately, bona fide within-segment recombination is thought to be extremely rare amongst commonly encountered pathogens with segmented negative sense single-stranded RNA genomes.
7.2.1 Considerations for Analysis
When it comes to analysing reassortant data, the most common approach is to infer the phylogenetic tree of each segment independently and at least initially compare them visually. This is entirely sufficient for rare and impactful reassortments (e.g., identifying reassortments giving rise to pandemic influenza A viruses). Tanglegrams are a visualization method frequently employed to make this visual comparison easier. In a tanglegram, typically two segment trees will be plotted with their tips facing each other and the tips that belong to the same genome connected with lines (Figure 7.2). The extent to which these lines criss-cross each other is taken as a proxy for the extent of reassortment. Keep in mind however that the y-axis position of any given tip in a tree is meaningless and that all of the relevant information (including the presence/absence of reassortment) is provided by the hierarchical nesting of the clades. It is possible to get phylogenetically incompatible trees that exhibit no criss-crossing of tanglegram lines because of how the clades are oriented! An extension of tanglegrams is called a tangled chain where multiple segment trees are typically plotted facing one way with tips belonging to the same genome connected using lines (Figure 7.3).
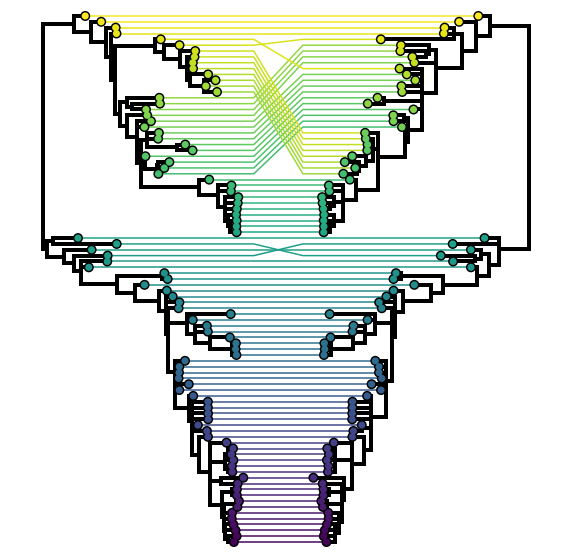
Figure 7.2: A traditional tanglegram. Tips belonging to the same genome are connected between a pair of trees inferred from different segments, in this case PB1 and PB2 segments of human influenza B viruses. For the most part the two trees resemble each other closely with the exception of a clade (yellow–green) at the top that has reassorted in the right tree with respect to the left.
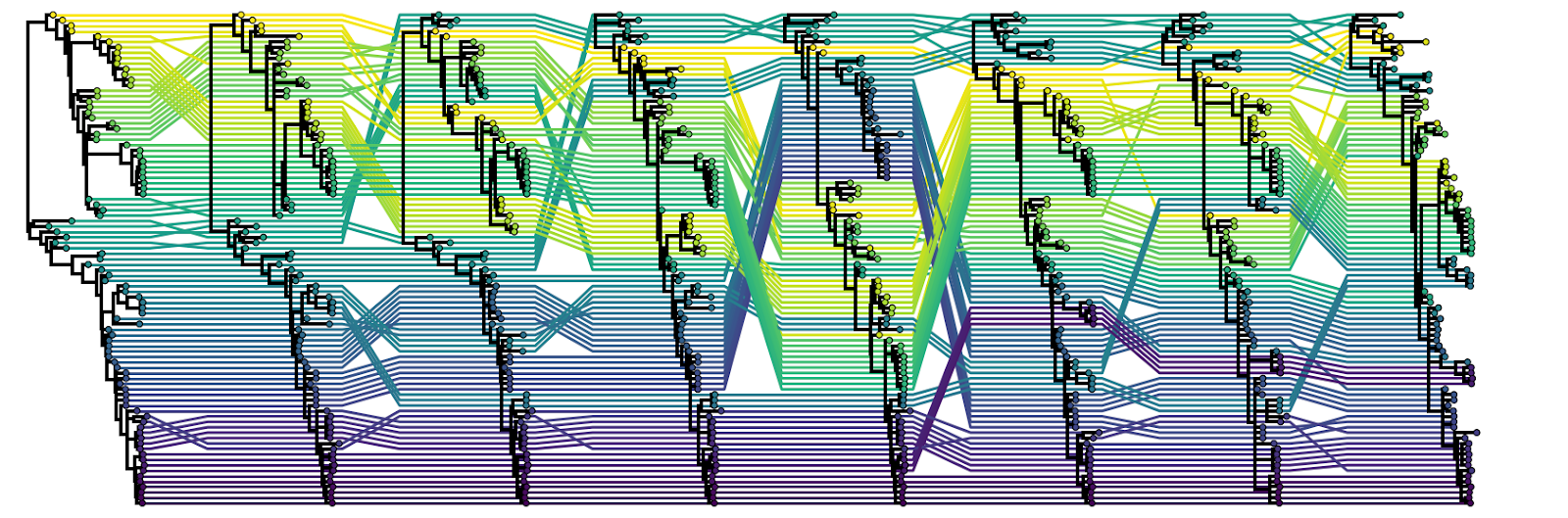
Figure 7.3: An example of a tangled chain. Here, all eight segment trees that comprise the human influenza B virus genome are plotted facing the same way and tips belonging to the same genome connected with lines.
Rigorous methods for reassortment inference exist and vary in terms of their computational demands and performance. GiRaF24 is an older method that infers reassortments from posterior distributions of segment trees inferred using Bayesian methods and reports clades that are reassortant and which segments were involved. TreeKnit25 is a recently developed method that takes in individual segment trees and reconstructs an ancestral recombination graph (ARG), a data structure that in addition to familiar splitting events also includes merging events that indicate reassortments and show what reassorting segments are most closely related and which corresponding segments were overwritten in the recipient genome. Another similar method that is particularly robust but intensive is the CoalRe26 package for BEAST227. It is intended for molecular clock analyses and in addition to recovering the posterior distribution of ARGs also returns the embeddings of each segment in ARGs, that is, the inferred time tree of each segment. An extension to the CoalRe method that can also reconstruct discrete character states (e.g., locations) is called SCORE.
7.3 Hypermutation
Hypermutation is not intentionally a method by which viruses generate genetic diversity; it is traditionally a host-defense mechanism, whereby the hypermutation should hopefully change the sequence so greatly as to render the progeny virion non-viable. However, hypermutation doesn’t always kill off a virus, and we can observe tracts of hypermutation in genomic datasets for certain pathogens. We will therefore discuss how to identify hypermutated regions of viral genomes, and how to handle those portions of the sequences when performing genomic epidemiological analysis.
There are a few eukaryotic enzymes that appear to be used by the host to attack viral genomes within cells. The most famous examples include APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide) and ADAR (adenosine deaminase acting on RNA), both of which sometimes leave distinct patterns of changes in viral genomes that survive encounters with these proteins.
APOBECs are the more diverse of the two and carry out cytidine deamination on RNA and DNA – they turn cytosines into uracils (RNA analog of thymine) such that a C:G pair becomes a U:G pair. The U:G pair is a mismatch, which will generally be repaired to a matched T:A pair, resulting in a C to T mutation.
There are many different APOBECs that are classified with numbers and letters (e.g., APOBEC1, APOBEC3A, etc.) whose functions are varied and potentially not understood fully at this time, and range from an antiretroviral role to aiding affinity maturation of B-cell receptors.
The function of ADARs, though somewhat clearer, is also not fully understood. They target double-stranded RNA (dsRNA) which is usually a very strong indicator of an RNA virus infection (cellular RNAs typically exist as either a DNA-RNA hybrid during transcription or single-stranded mRNA, tRNA, or rRNA otherwise). The replication of RNA virus genomes produces an intermediate step that is double-stranded RNA. An ADAR enzyme recognizing this dsRNA may deaminate adenosines into inosines which behave like guanines (i.e., pairing with cytosine), resulting in A to G mutations.
When looking for putative host-driven hypermutation patterns in viral genomes, you should be aware of the nucleotide context that the enzyme in question prefers. For example, APOBEC3 has a preference for changing TC to TT, that is, the edits of C-to-T are biased if there’s a preceding T. As an additional point of consideration, edits may appear in two different ways depending on whether edits are occurring on the sense or antisense strand. For example, if we were looking for ADAR edits, we should look for both A to G and T to C mutations since dsRNA can be approached (and edited) from either side.
As a concrete example, during the West African Ebola virus epidemic there were a number of Ebola virus genomes that carried stretches of tens to a bit more than a hundred nucleotides with clustered T to C mutations but not A to G (Figure 7.4). The current thinking goes that ADAR preferentially edits As into Gs on the sense strand (as opposed to the antisense, that is, the reverse complement of the genome) of negative sense single-stranded RNA viruses. Since bioinformatically we overwhelmingly work with RNA virus genomes pointing the “conventional” direction (5’ to 3’ with genes running left-to-right) we observe these as stretches of T->C mutations.

Figure 7.4: An example of a strong ADAR editing signal in the Ebola virus genome from the West African epidemic. The upper sequence (Makona-Gueckedou-C05) is an early sequence from the epidemic while the bottom one (J0169) is from an outbreak in Freetown later in the epidemic and bears a high number of T/U->C mutations in a very short stretch. There are about as many mutations in this small stretch as there are other non-clustered mutations differentiating these genomes.
If you observe putative tracts of hypermutation and you plan to run molecular clock analyses on these sequences, you should seriously consider masking the sites of putative editing. Any molecular clock models the slow trickle of mutations that are introduced by the viral polymerase and that manage to survive by not being sufficiently deleterious. A sudden influx of lots of mutations (in Figure 7.4 it’s roughly doubling the number of mutations there were before) the molecular clock can only assume that the overall molecular clock rate is much higher and therefore the entire tree is much younger. This is less of a problem for more sophisticated clock models, like relaxed uncorrelated clocks, but should still be addressed head-on if there are strong suspicions that questionable mutations came about via anything other than the viral polymerase making errors.
Note that we will not always have the luxury of neatly defined stretches of hypermutation. During the mpox (previously called monkeypox) outbreak of 2022, there were strong indications that the clade causing the outbreak had at some point experienced APOBEC3 editing albeit without a spatial clustering as extreme as the Ebola virus example in Figure 7.4.
7.3.1 Handling Variable Mutation Rates with Relaxed Molecular Clocks
In Chapter 2, Section 2.3 we discussed molecular clocks, and how you could estimate the evolutionary rate by fitting a linear regression line through a root-to-tip plot. This type of clock is referred to as a strict clock. It is considered a strict clock because it fits the same evolutionary rate to all branches in the tree.
The idea that all branches in the tree evolve at the same rate is a simplifying assumption that lowers the parameterization of the analysis and generally makes computational analyses run faster22,27. Strict clocks often work well, but there may be times when you want to capture evolutionary rate heterogeneity across the tree. To do so, you need to estimate branch-specific evolutionary rates, and we can do this using relaxed clocks. One common method for estimating a relaxed molecular clock is using BEAST or BEAST2. While there are multiple ways to specify the relaxed clock, most commonly we consider the rate heterogeneity to be log-normally distributed and uncorrelated.
What does this mean? Firstly, branch-specific evolutionary rates are drawn from a log-normal distribution, which is a probability distribution that looks like a normal distribution in log-space. The draws are considered uncorrelated because each branch gets an independent draw regardless of where the branches are located in the tree, as opposed to forcing parent-offspring branches to have some correlation in their rates. During analysis, the parameters of the log-normal distribution, the mean and standard deviation, that best fit the sequence data at hand are estimated. While you can look at the branch-specific rates, the mean rate across the entire tree is computed by summing all the mutations that must have happened across the tree (computed as the evolutionary rate for each branch multiplied by the length of that branch) and dividing that sum by the sum of all branch lengths in the tree.
Why might you want to use a relaxed clock? Firstly, using a relaxed clock prevents branches with more or fewer mutations than the average from having an undue influence over the molecular clock rate across the entire tree. Essentially, it prevents the average evolutionary rate from being unduly influenced by outliers (such as might occur if you haven’t masked tracts of hypermutation). Secondly, sometimes branch-specific rate variation is one of the specific quantities that you would like to measure. For example, when investigating cases of sexual transmission of Ebola virus disease from survivors, a common indicator that such transmission has occurred is that the recipient viral infection looks genetically quite similar to the strain that the infector was infected with, even when many months have passed. On a tree estimated under a relaxed clock, this relative lack of genetic divergence given the amount of time that has elapsed between the infector’s infection and the infectee’s infection will show up as a much slower branch-specific evolutionary rate. For examples, see Mbala–Kingebeni and colleagues’ investigation of an Ebola virus disease relapse infection14, or Rambaut, McCrone, and Baele’s (2019) tutorial on using relaxed clocks in BEAST to estimate Ebola virus rate heterogeneity (https://beast.community/ebov_local_clocks.html).
7.4 Diversity of RNA Virus Genome Organizations
There will be many cases where you will follow standards of analysis set by the pathogen’s research field. However, if you’re ever in a position where you encounter a new pathogen or decide to venture outside your comfort zone and explore other pathogens, you might encounter some evolutionary solutions viruses have come up with that may affect the way you carry out your analyses. You should be aware of these unique edge cases and consider how your methods might be affected by such sequences. The following is by no means an exhaustive list of considerations but one that might give you an appreciation for what to be on the lookout for.
7.4.1 Ambisense
Though for the most part many RNA virus genomes code their genes in the same direction (i.e., RNA polarity – 5’ to 3’ or 3’ to 5’), there are some groups that vary in this regard. One extreme example (and not relevant for human public health at the time of writing) are narnaviruses, some of whose members contain two open reading frames running in opposite directions that overlap across the entirety of their segment(s). Only one of these is thought to be functional. Human-relevant viruses with ambisense genomes include arenaviruses (e.g., Lassa and Machupo viruses) where each segment contains a reverse-complementary loop sitting between a pair of genes each “pointing” towards the loop. If you are working at the nucleotide level, further analyses might not be affected, but you should keep these nuances in mind if you decide to work at the amino acid level and are not using programs that support annotations and their automated extraction/translation from nucleotide sequences.
7.4.2 Reverse Complementarity
At some point you may encounter viruses that use RNA structures to complete their life cycles. Hepatitis delta virus, the causative agent of Hepatitis D, is a satellite virus that can accompany hepatitis B virus infection. Hepatitis delta virus has a circular single-stranded RNA genome which is highly self-complementary and so adopts a partially double-stranded RNA structure. Another example of reverse-complementarity in RNA viruses includes segment ends that are used for genome packaging.
The easiest way to check whether such sub-sequences exist in a given genome/segment is to run self-similarity dot plots. There are a number of software packages that can do that for you (e.g., dotmatcher). They will plot a pair of sequences (can be same or different), one along the x-axis and one along the y-axis, and put a dot for every nucleotide that matches. If plotting a sequence against itself, you should see a diagonal, one-to-one, line running through the plot (since the sequence is identical to itself at each position). Any off-diagonal elements indicate repeats, and if the method allows for complementarity, any offset diagonal lines oriented the opposite to the diagonal one-to-one line will indicate a region that is reverse complementary to another region.
7.4.3 Splicing
Sometimes viruses may process their mRNAs via splicing, which can present a challenge for interpreting sequencing data and/or evolutionary analyses. Influenza A viruses are famous examples of this where transcripts of at least two segments (matrix and non-structural) are spliced before being translated into at least four proteins with distinct functions. Splicing may be suspected in RNA virus genomes if there appear to be regions with excess non-coding space. For many RNA viruses, such genomic space is a premium resource and so must be explained. You can strengthen your suspicions by looking at whether long open reading frames offset from the “main gene” exist nearby, and get evidence of splicing by sequencing infections to a great depth since occasionally you will be able to catch both the genomic unspliced and spliced transcript RNAs. Note that splicing happens in the eukaryotic nucleus and thus might not apply to the virus you’re working with.
