Mind-blowing science: ‘Star Wars-style’ holograms to communicate with the brain
About 20 years ago, neuroscientists, recording from electrodes implanted in the medial temporal lobe, identified human brain cells that respond only to photos...
From launching a new AI challenge to speed up annotation to helping researchers more easily process imaging data, the CZ Imaging Institute is making progress toward its mission to develop technology that pushes the boundaries of what we can see and measure in cells.
Cryo-electron tomography (cryoET) gives us powerful information on the inner workings of the cell at near atomic resolution. However, the analysis of the data, known as tomograms, remains a significant bottleneck to discovery as it often requires manual labeling and annotation. The CryoET Data Portal, created by the CZ Imaging Institute, is an open access data repository that contains about 16,000 annotated cryoET datasets in standardized formats.
A new correspondence in Nature Methods from the Chan Zuckerberg Institute for Advanced Biological Imaging (CZ Imaging Institute) gives insight into how scientists and ML researchers created the CryoET Data Portal, as well as goals and next steps in support of the portal. Read the full correspondence.
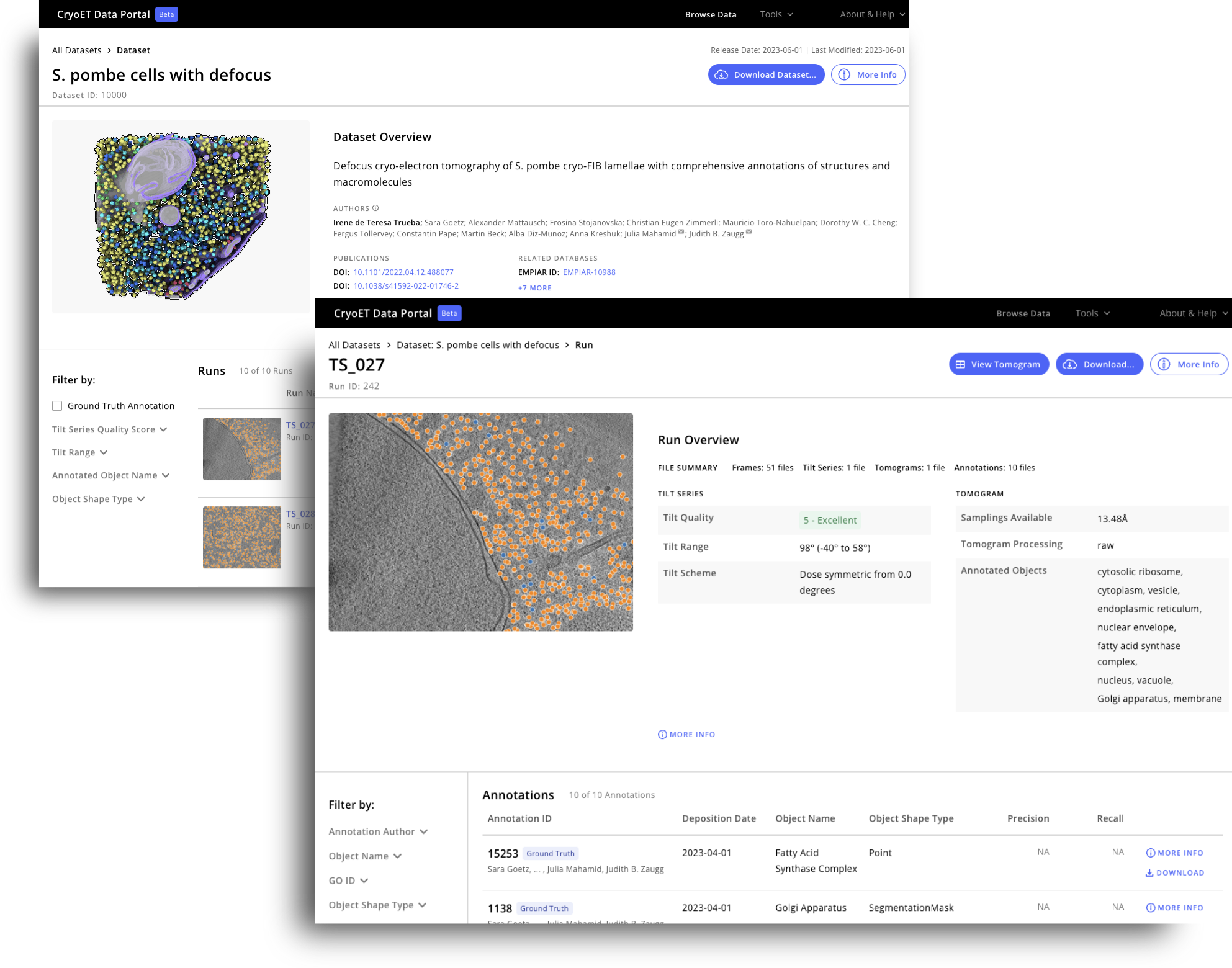
CryoET can now provide thousands of 3D images—called tomograms—in less than a week. But to realize the full potential of this technology, biological objects like proteins and membranes need to be identified in the tomograms through annotation. Since manual labeling can take months or even years, there is an urgent and immediate need for machine learning algorithms that can robustly annotate particles of different shapes, sizes and abundances within cells in order to unlock the discoveries currently hidden in thousands of existing tomograms.
The CZ Imaging Institute is hosting an international competition to advance our understanding of cell biology by developing machine-learning algorithms that can annotate biological particles in 3D images of cells captured by cryoET. These algorithms should be able to perform robust annotation of particles of variable shapes and sizes within the hundreds of 3D images in the competition dataset after being trained on a limited set of available reference annotations from the same dataset. To reduce the onboarding time for competitors, an extensive set of example notebooks is being provided.
The competition will run until February 5, 2025, and prizes will be awarded. Interested in entering the competition, or have peers who might be? Visit cryoetdataportal.czscience.com/competition for more information.
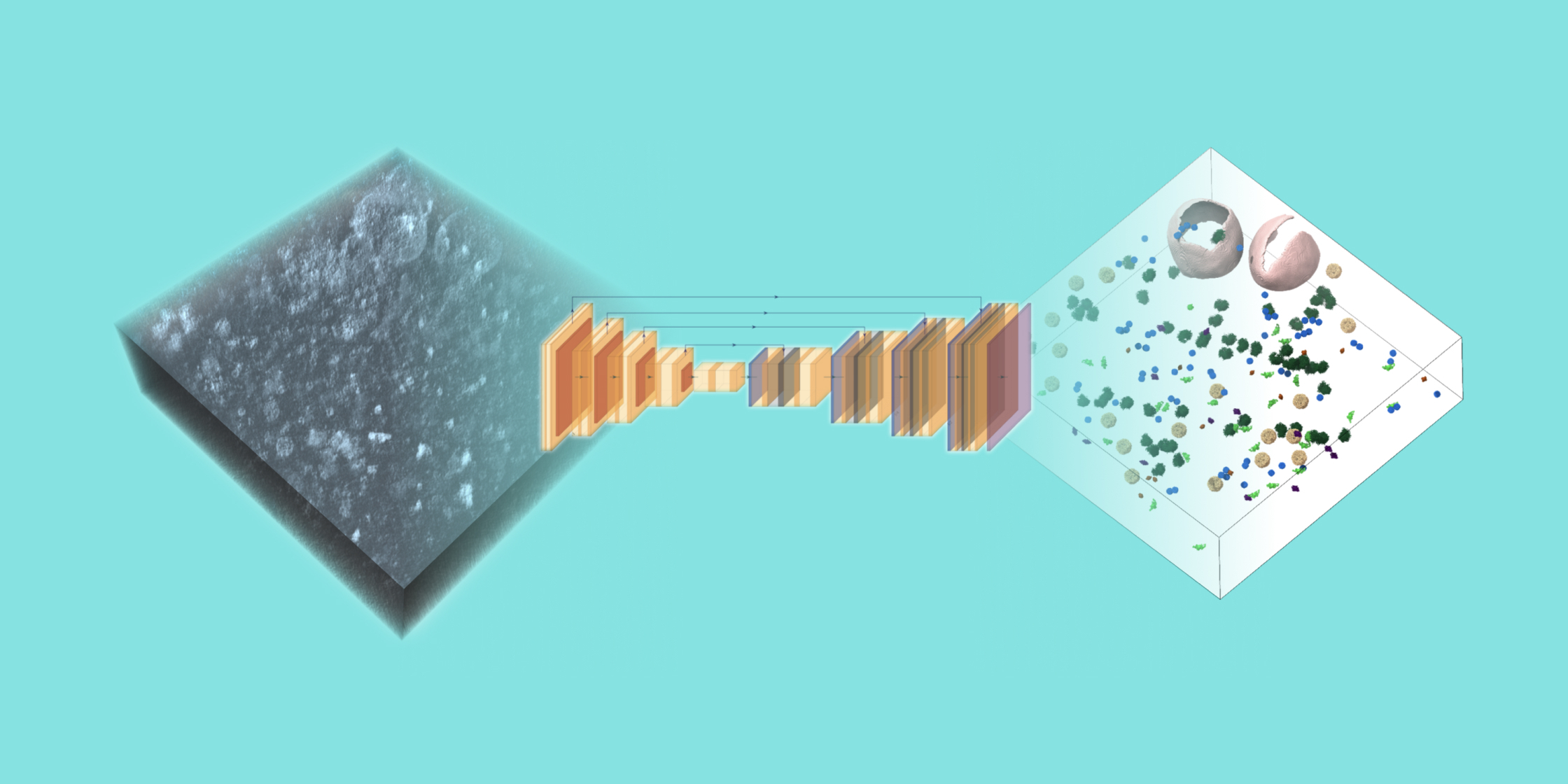
One of the greatest difficulties with processing cryoET data is the challenge of scale: scientists often need to locate thousands of copies of a molecule of interest across hundreds of tomograms, i.e. three-dimensional maps of an imaged cell, to reveal its structure in situ.
Earlier this year, the CZ Imaging Institute released AreTomo3, a software package that provides a fully automated and accelerated pipeline to transform the raw images collected on an electron microscope into three-dimensional tomograms that reveal the molecular architecture of cells. The platform’s beginner-friendly interface reduces barriers for new users to process their data and facilitates seamless integration with software packages from across the cryoET community. AreTomo3’s focus on accuracy and speed enables users to immediately begin downstream data processing tasks even while data collection is ongoing. Tools like AreTomo3 are critical to advance the pace of scientific discovery by automating and simplifying processes that might otherwise take significant amounts of manual labor and time.
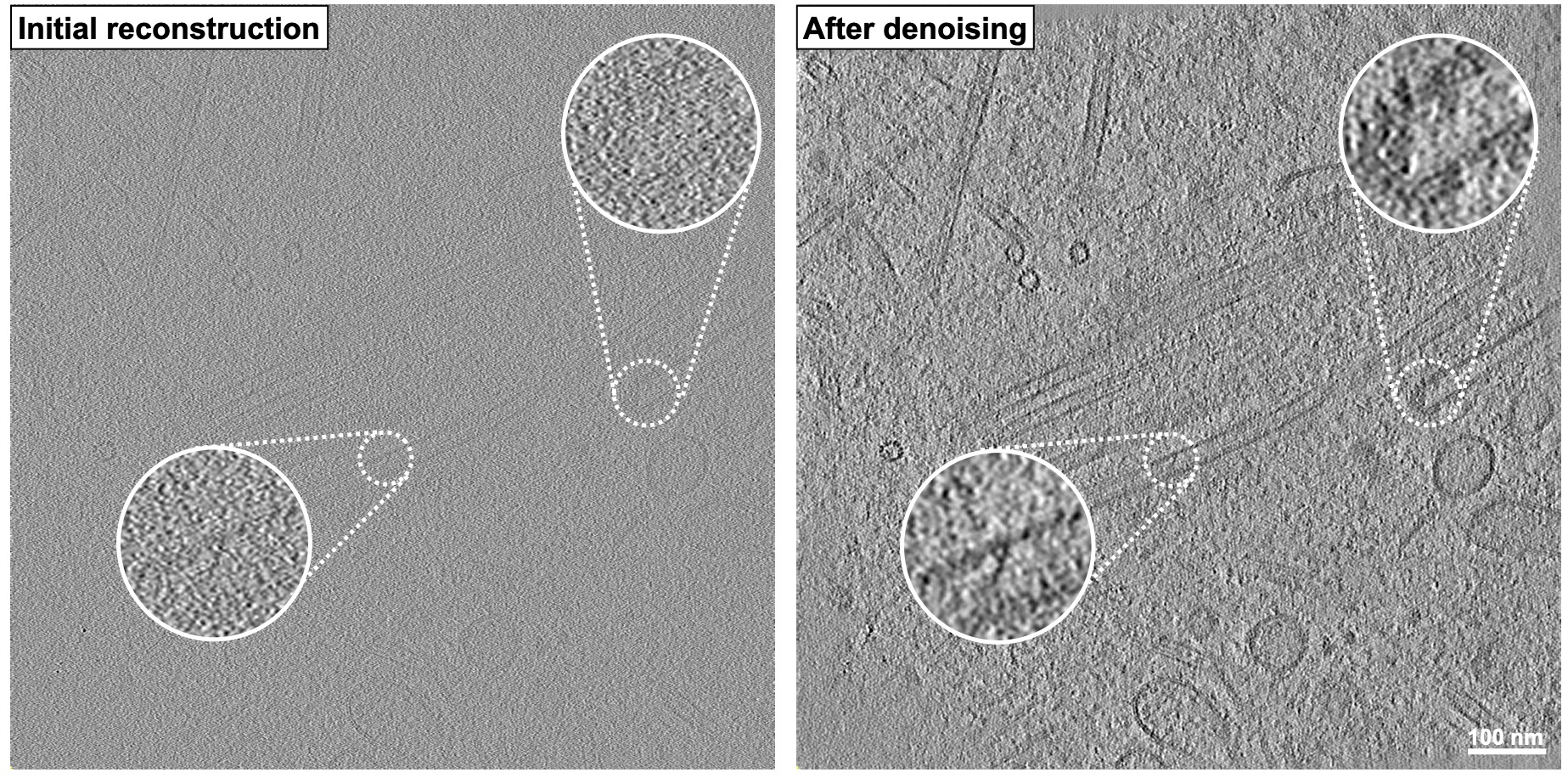
The team recently released a new version of AreTomo3, which builds on the original release by providing multiple entry points to accelerate data reprocessing, sample thickness measurements to improve alignment accuracy, and robust corrections for the microscope’s contrast transfer function to improve downstream denoising, among other features. Learn more about AreTomo3 and other biological imaging tools created by the CZ Imaging Institute.
To keep up to date with the latest from the CZ Imaging Institute, sign up for our mailing list.
About 20 years ago, neuroscientists, recording from electrodes implanted in the medial temporal lobe, identified human brain cells that respond only to photos...
Learn More
A ‘Google Earth’ of embryology, CZ Biohub San Francisco's zebrafish cell atlas brings a new vision to developmental biology
Learn More
Using frogs as a model organism, CZ Biohub SF Investigator Helen Willsey is revealing surprising connections between autism and hair-like structures on the...
Learn More
Stay up-to-date on the latest news, publications, competitions, and stories from CZ Biohub.
Cookies and JavaScript are required to access this form.