Our Research
Ribosomes are intricate protein-making machines essential for all life on Earth. We explore ways of modulating their function to reduce the production of pathogenic proteins, with a particular interest in viral and genetic triggers of neurodegeneration.
What do ribosomes need in order to synthesize specific pathogenic proteins? And how can we exploit such dependencies to selectively inhibit the process, and move towards novel protein-lowering therapies?
To produce new proteins, ribosomes rely on a highly complex, modular, and dynamic network of interactors. These include components that bind the ribosome, mRNA template, and nascent polypeptide products. Together, they remodel RNA structure, govern translation kinetics, fold and modify nascent chains, and target newly synthesized proteins to specific subcellular compartments.
Using a combination of molecular biology, mass spectrometry, and CRISPR screens in human iPSC-derived neurons, we explore how this network is remodeled in response to disease perturbations, including acute infection with neurotropic viruses or persistent expression of mutant proteins that cause neurodegeneration. We harness this knowledge to design targeted interventions that could alter the course of disease by lowering the levels of pathogenic proteins.
Projects
Host-directed therapeutics for antiviral interventions
Most antiviral drugs target viral enzymes. Because viruses mutate rapidly, such interventions often carry a high risk of resistance. An alternative approach is to target host components that are both essential for the virus lifecycle and dispensable for short-term host survival. Our work aims to discover and characterize host factors that facilitate production of mature viral proteins and can be targeted using small-molecule inhibitors for broad-spectrum, low-resistance antiviral interventions.
Protein-lowering strategies in neurodegenerative disorders
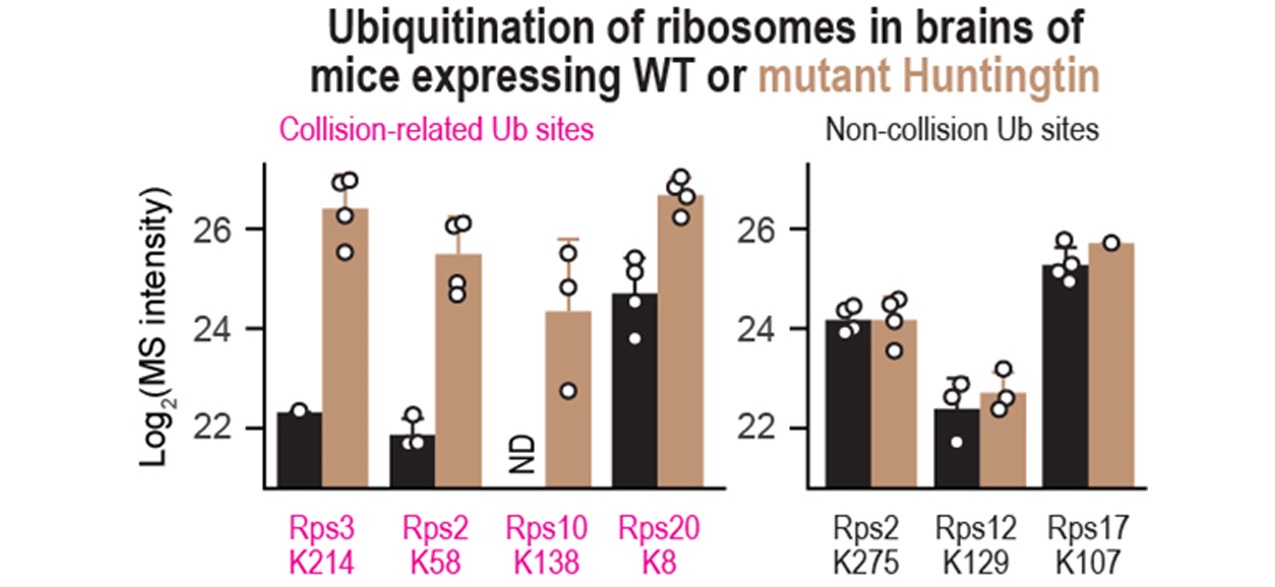
A wide range of human pathologies are associated with changes in ribosome function. Furthermore, many neurodegenerative disorders involve the production of mutant proteins that form toxic aggregates, leading to cell death. By analyzing ribosome-associated networks and translation kinetics, we aim to improve our understanding of disease etiology and identify targets for selective lowering of such toxic proteins.
Tools to explore ribosome function
Recent developments in high-throughput technologies have revolutionized our understanding of protein synthesis. Our work aims to expand the available toolkit by developing molecular reporters and proteomic assays to monitor ribosome function and translational output. We are applying these methods to identify neuron-specific regulators of protein synthesis and explore how translation errors are involved in human disease.