How technology is democratizing genetic research for rare diseases
Discover how a new genetic prevalence estimate calculator can support advancements and new insights for rare disease research.
Farmers and meatpackers once streamed into Chicago’s bustling wholesale district at Fulton Market, arriving to sell produce, hay, and cattle. Now, over 100 years later, on a beautiful fall day in late September, scientists stream into this bustling innovation district, arriving for a two-day conference on next-generation technologies in the life sciences hosted by the Chan Zuckerberg Biohub Chicago.
CZ Biohub Chicago launched in 2023 with the ambitious goal of engineering novel technology platforms to understand, and even someday control, one of the most complex and important phenomena in the human body — inflammation. An estimated 50% of all deaths are attributed to inflammation-related diseases, including cancer, heart disease, Alzheimer’s, and infectious diseases.
“We hope to ultimately discover and define the rules that control inflammation up and down in the human body,” said Shana Kelley, President of CZ Biohub Chicago and professor of chemistry and bioengineering at Northwestern University, during opening remarks at the conference. “Through the data we collect and the technology we develop, we can gain the ability to modulate master regulators of inflammation, and eventually have new therapeutic strategies in hand to intervene when inflammation gets going.”
A critical step in that effort is the close cooperation of previously disparate fields, including cell biology, engineering, systems biology, physics, and more. As a scientific kickoff for CZ Biohub Chicago, Kelley and the Biohub leadership invited top scientists from across these fields to come together in person, share insights on cutting-edge sequencing, imaging, and bioengineering platforms, and challenge each other to probe and question living systems from new perspectives.
“We will have an unwavering focus for the next 10 to 15 years on creating the tools that are going to help us make an impact on our goal — to cure, prevent, and manage all disease by the end of this century,” said Amy Herr, Vice President of the CZ Biohub Network and bioengineer at the University of California, Berkeley, to the gathered crowd. “We are thinking about what tools are going to be needed next.”
With our feet in the present and eyes on the future, here are our top four insights from this seminal meeting at the intersection of biology and engineering:
In 2012, two biologists won the Nobel Prize in Chemistry for identifying the gene sequence, structure, and function of a single family of cell receptor proteins. In 2024, the Nobel Prize in Chemistry was awarded to a biochemist, David Baker, and two computer scientists, Demis Hassabis and John Jumper, for developing computational tools that can predict and design protein structures — a feat thought near impossible. With the use of AI, Hassabis and Jumper made it possible to predict the complex structures of virtually all 200 million known proteins in nature.
That dramatic shift from single molecule wet lab work to complex bioinformatic analysis and deep learning tools has never been more apparent in biology. “The life sciences are becoming data sciences,” said Rashid Bashir, member of CZ Biohub Chicago University Advisory Committee and professor of Bioengineering at the University of Illinois Urbana-Champaign, during introductory remarks for a panel on trends in biomedical innovation. “Ten years from now, data science will be part of all the science done in the life sciences.”
Today, academic labs, government institutes, research hospitals, and research organizations like CZ Biohub Network collect, aggregate, and curate terabytes of high-resolution biological information about genes, proteins, cell-to-cell interactions, tissue function, and the epidemiology of human disease. Yet, as Bashir and other speakers emphasized throughout the conference, more data isn’t always better data: The type, quality, and breadth of data in biological experiments matter. This includes not only sequencing data but 3D imaging and multimodal data sets, many of which were discussed during the conference.

For example, Neil Kelleher, CZ Biohub Chicago Investigator and professor of Chemistry at Northwestern University demonstrated how detailed, single-cell data can help capture the complexity of a human cell. Using single-molecule mass spectrometry — a powerful technique to measure individual proteins, lipids, and metabolites in single cells — his team and collaborators at the Human Proteoform Project are mapping all human proteins, including their numerous molecular forms called proteoforms, the way the Human Genome Project mapped genes.
Some researchers have been skeptical that all the proteoforms in a single cell could be measured, but with quality data from single-molecule mass spectrometry, and AI to help analyze that data, “the proteome is able to be measured and sequenced in our lifetimes,” said Kelleher.
In this example and many others presented at the conference, AI and machine learning tools were key elements in analyzing and understanding large datasets. “We used to be at the threshold of the AI revolution. Now we are in it,” said Loic Royer, Senior Group Leader and Director of Imaging AI at CZ Biohub San Francisco.
Royer shared a glimpse inside a time-resolved, multidimensional atlas of vertebrate development, which is being developed at CZ Biohub SF and uses zebrafish as a model organism. Building Zebrahub required deep learning-based image processing and analysis algorithms, and today an AI-integrated widget called Omega that Royer built allows users to explore the resource through natural language commands. “These models are the currently worst they are ever going to be,” said Royer with a smile. “They’re only going to get smarter.”
In 1992, scientists sequenced the RNA transcripts of a small number of genes in a single cell. Seventeen years later, a team sequenced the whole transcriptome — that is, the complete collection of RNA transcripts — for single mouse germ cells. Jump ahead another 15 years, and single-cell transcriptomics, as well as single-cell genomics, proteomics, metabolomics, and more, are common, even routine technologies.
Omics tools are now faster, cheaper, and more accurate than they’ve ever been. “After 30 years of technology development, these tools have reached their full potential,” said Steve Quake, CZI’s Head of Science. “Now, it’s time to see what they can do.”
At the conference, Quake presented the latest developments of the Tabula Sapiens Consortium at CZ Biohub San Francisco, which has built a massive digital atlas of transcriptomes from nearly 500,000 single cells from 24 human tissues and organs. With that tool, researchers can now study human transcription factors with unprecedented scope. As an example, Quake described the discovery of transcription factors that are ubiquitous in human cells but were previously uncharacterized.
Large-scale single-cell omics are no longer just for hypothesis generation, but to learn deeply about human biology, added Rong Fan, a bioengineer at Yale University. Fan presented work on massively parallel single-cell gene expression profiles to figure out why some people have durable responses to CAR-T cell therapies while others don’t. Next, his team is combining image-based and sequencing-based technologies to create “multi-omics” maps that AI can help explore and understand.
Envisioning and building technologies that will enable the next leaps in omics research is part of CZ Biohub Chicago’s commitment to high-risk, high-reward research. “We want to be working outside the comfort zone of traditional research and pushing ourselves to take on approaches that are too risky and too far out to fit into existing funding paradigms,” said Kelley.
Throughout two days of sessions and panels, one theme promptly kept resurfacing: Static datasets, such as the transcriptome of a single cell at a single time point, do not give a full picture of dynamic, living systems. To truly understand complex biological phenomena, experiments need to take into account the changes that occur in and between cells and tissues across time and space.
“Everybody is interested in spatial biology,” said Savas Tay, a CZ Biohub Chicago Investigator and a professor of molecular engineering at the University of Chicago. “When we rely on dissociated cells, we are losing a lot of information and context.”

For example, Emma Lundberg and colleagues at Stanford University found that 55% of human proteins localize to multiple compartments in cells, indicating that many proteins are involved in multiple biological processes, and some move from space to space. “With this in mind, I’m going to argue it’s hard to truly understand cell function or model cells if we don’t do that with spatial resolution,” said Lundberg, also a CZ Biohub SF Investigator, during a presentation.
She and collaborators are building the Multi-Scale Integrated Cell (MuSIC) Project, the first systematic map of human cell structure that integrates both spatial and temporal protein data that is, the activity of proteins across time and space in a cell. Their effort will rely on machine learning to interpret and integrate imaging and mass spectrometry data. Once complete, they hope to perturb the model to identify novel functions of proteins and the consequences of metabolic changes in a human cell and therefore discover new drug targets for metabolic conditions.
At CZ Biohub Chicago, group leader Daniel Wang and his team are pioneering a new tool for spatiotemporal analysis that uses microneedle tissue sampling to collect biomolecules from numerous locations in a tissue simultaneously. “Inflammation is complex in space and time,” said Wang. “We need new technologies that are spatially resolved and minimally invasive.”

Gathering and analyzing spatial and temporal data adds a layer of complexity to experiments that researchers are just beginning to grapple with. “As we are generating more data, there are basic challenges in integrating these data across scales and time,” said Gordana Vunjak-Novaković, a CZ Biohub New York Investigator and biomedical engineer at Columbia University, whose team develops organs-on-a-chip platforms for disease modeling and drug development. “How do we know we are measuring the right thing? And how simple is complex enough?”
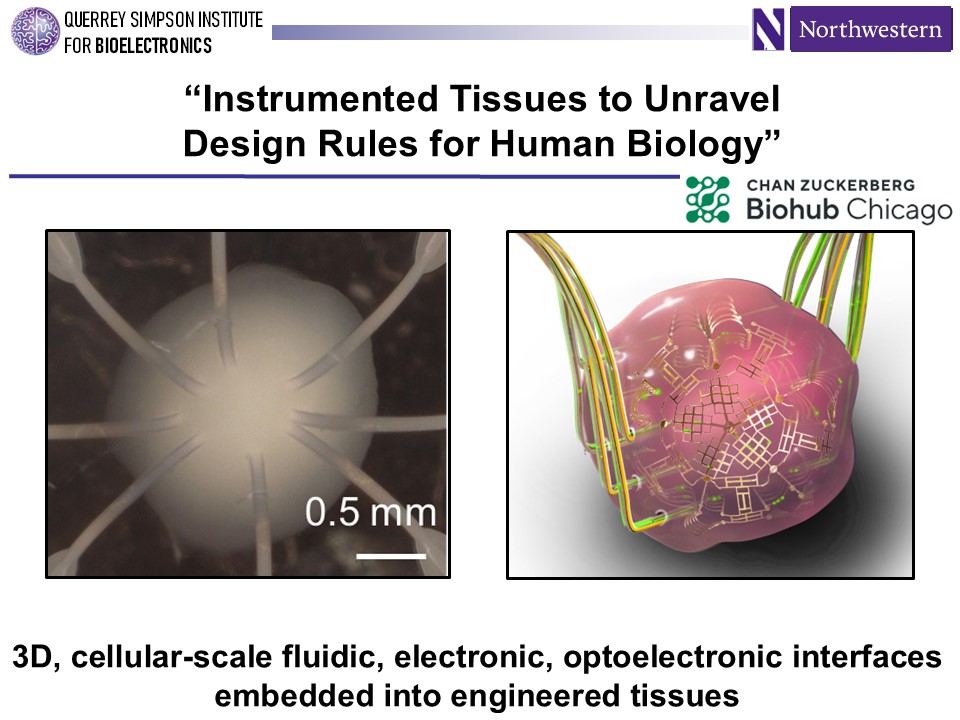
One way to know if the right thing is being measured is to embed electronics directly into living tissues to monitor, directly sample, and even control the tissue being studied.
In pursuit of “instrumented tissues,” researchers presented a range of new tools for sensing and directly measuring inflammation and other conditions within tissues. Chemist and materials scientist John Rogers of Northwestern University described 3D silicon structures that can be embedded with sensors and act as a scaffold to grow organoids. “We have the building blocks for embedding electronics into small-scale tissue structures,” said Rogers, a member of the planning committee for CZ Biohub Chicago.
Bashir shared videos of fabricated muscle strips, engineered with electronics and muscle cells that flex and walk when activated with light pulses from a microLED device. These cell-based soft robotics provide a way to build biological machines that can sense and process input signals. Alice Ting, a CZ Biohub San Francisco Investigator and professor of genetics at Stanford University, described modular, programmable receptors that can act as biosensors in cell membranes, reacting to stimuli in the outside environment by emitting a response, such as glowing or secreting an antibody.
“The cell is an incredible, programmable device,” concluded Mike Elowitz, a professor of biology at Caltech, in the final presentation of the conference. To what extent, he wondered aloud to the crowd of scientists, can we engineer tools to program and control these complex behaviors?
With AI, advanced omics, spatial and temporal data, and instrumented tissues all pushing the frontier of bioengineering, we can’t wait to find out.
Learn more about CZ Biohub Chicago’s mission to build state-of-the-art technologies to monitor and change immune activity in real time.
Discover how a new genetic prevalence estimate calculator can support advancements and new insights for rare disease research.
Learn More
Using frogs as a model organism, CZ Biohub SF Investigator Helen Willsey is revealing surprising connections between autism and hair-like structures on the...
Learn More
Scientists have been trying to understand how the cells in our body work since the invention of the microscope. Meet five scientists taking these efforts to...
Learn More
Stay up-to-date on the latest news, publications, competitions, and stories from CZ Biohub.
Cookies and JavaScript are required to access this form.