Family loss spurs quest for knowledge
For CZ Biohub San Francisco research associate Rodrigo Baltazar-Nuñez, science is personal
Modern imaging technologies have opened windows to see inside human cells at high enough resolution to observe individual proteins. This detailed view of the inner workings of cells can reveal unanswered questions about the architecture of a healthy cell, and the changes that may lead to disease. While technological advancements have cut down the capture and processing time, a major bottleneck in discovery remains in the annotation and analysis of these data-rich images.
Recently, CZI reached a new milestone towards this challenge when researchers at the Chan Zuckerberg Institute for Advanced Biological Imaging annotated membrane proteins in over 13,000 3D images added to the open access cryoET Data Portal. By applying new machine learning tools to the data and running analyses using large-scale computing, they were able to achieve this otherwise near impossible feat in just three and a half days.
Slices through a 3D tomogram of a human umbilical vein endothelial cell (HUVEC) grown on an EM grid from Dataset 10176. This exemplifies the challenge of annotating tomograms: cells are very crowded, tomograms have very poor contrast and are anisotropic, and individual particles to annotate are sparse. The scale bar is 200 nm and the total field of view size is 2000×2000 nm. Dataset (Credit: Cora Woodward; Grant Jensen)
Cryo-electron tomography (cryoET) is an imaging technique that enables 3D visualization of the cell at sub-nanometer resolution but, unlike other high-resolution imaging techniques, the cryogenic (frozen) condition preserves cellular architecture so this detailed view includes protein structures in their natural biological context. Three-dimensional tomograms can be generated from hundreds of images of a thin slice through a cell, taken while tilting the specimen in multiple directions. A given tomogram is typically only about 200 nanometers thick—approximately five hundred times thinner than a sheet of paper—yet packed with information about the structures of the cellular machinery driving health and disease.
Acquiring 3D tomograms can take several days, but annotation is necessary to turn these data into useful insights. Annotation involves tracing sparse particles of subcellular structures through many layers of low contrast, two-dimensional images. It’s similarly painstaking to tracking a single pixel in a black and white movie—by hand. Doing this manually on a set of tomograms can take months, and on the total volume of data that already exists is impractical.

To accelerate this process, the CZI and the CZ Imaging Institute created the cryoET Data Portal to give biologists and developers open access to high-quality, standardized, annotated data they can readily use to retrain or develop new annotation models and algorithms. All tomograms in the cryoET Data Portal include rich standardized metadata such as data tree structure and naming conventions. The CZ Imaging Institute is working with developers at EMPIAR to ensure the portal is compatible with their resource, and CZI recently funded a workshop with EMPIAR’s founding group, the European Bioinformatics Institute (EBI), to discuss metadata standards.
Until recently, the portal had 720 tomograms with annotations submitted. However, when Grant Jensen contributed 12,597 unannotated tomograms to the portal, CZ Imaging Institute imaging scientist Utz Ermel saw it as an opportunity to put a new deep learning pipeline called MemBrain to the test.
After the submitted tomograms were ingested into the data portal through a pipeline developed by engineers at CZI in collaboration with CZ Imaging Institute, Utz made some modifications to the pre-trained MemBrain model and applied it to the new tomograms.

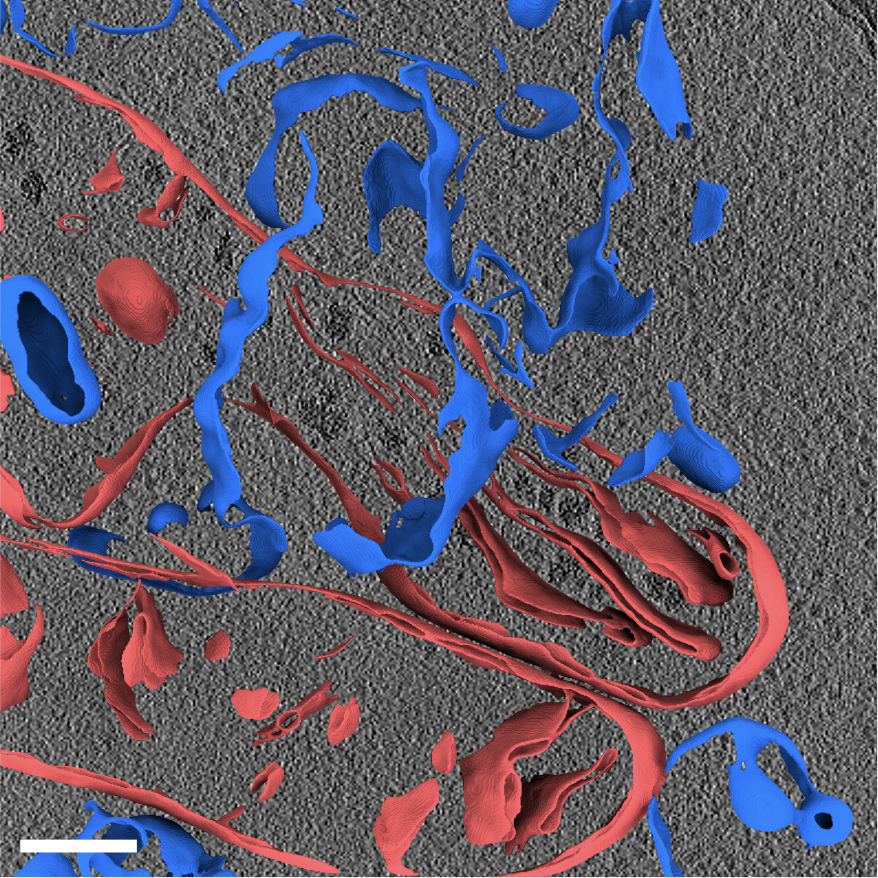
In just three and a half days, the membrane segmentation algorithm Utz adapted was able to annotate membranes of 13,275 tomograms. At the same time, Utz had been working with Robert Kiewisz and Tristan Bepler from the Simons Machine Learning Center at New York Structural Biology Center, who submitted their own annotations of these tomograms using a different segmentation algorithm called TARDIS. This marks a milestone in open collaboration between independent groups through the portal since the website launched in December 2023.
With two different approaches to membrane annotation now available on the same set of data in the portal, this provides a ripe opportunity for collaboration to improve membrane segmentation methodology and adapt them to additional cell structures. While membranes are a great starting place because of their uniformity across species, scientists at the CZ Imaging Institute are keen to work on algorithms to identify individual macromolecules, especially those associated with membranes as these tend to be the target for the majority of therapeutics and understanding them in the context of cells is important. They are also aiming to annotate filamentous proteins, including the segmentation of microtubules and actin. From there, they can tackle increasingly challenging proteins, including those with more inconsistent conformations.
36 exemplary membrane segmentations for Dataset 10007, FIB-milled thin sections of Baker’s Yeast, containing 263 tomograms in total. The scale bar is 200 nm. Dataset Authors: Ramya Rangan; Sagar Khavnekar; Adam Lerer; Jake Johnston; Ron Kelley; Martin Obr; Abhay Kotecha; Ellen D. Zhong
Currently, the portal contains 15,138 tomograms from 298 datasets along with annotations, contributed by the groups of Julia Mahamid, Jürgen Plitzko, David Agard, John Briggs, Abhay Kotecha, Ellen Zhong, Ben Engel, Danielle Grotjahn, Grant Jensen, Robert Kiewisz and Tristan Bepler. All tomograms, including annotations, are published with a CC0 license, meaning they are available in the public domain for other researchers to freely download, share, and use for their work.
Researchers at CZ Imaging Institute are committed to standardizing metadata and annotations, and will continue running new algorithms on the portal data as they become available and sharing the findings with the community. While the portal is only a year old, with a growing number of contributors, this collaborative approach will improve our understanding of how all of the components of a cell come together—in different cells and during different states of health, disease and age.
For CZ Biohub San Francisco research associate Rodrigo Baltazar-Nuñez, science is personal
Learn More
Using frogs as a model organism, CZ Biohub SF Investigator Helen Willsey is revealing surprising connections between autism and hair-like structures on the...
Learn More
CZ Biohub SF-backed NextGen initiative succeeds in diversifying faculty hiring in biological and biomedical sciences at major Bay Area universities
Learn More
Stay up-to-date on the latest news, publications, competitions, and stories from CZ Biohub.
Cookies and JavaScript are required to access this form.