When early cartographers undertook perilous expeditions to map unknown corners of the world with sextants, compasses, and hand-drawn diagrams, it’s unlikely they imagined that someday anyone with an internet connection would have access to a seamless view of the entire planet from the comfort of their own home. Today, pioneering scientists are working to create a similar experience for a much tinier, but no less important domain: developing embryos. The goal is to track and map the behavior of each and every cell working together to create an adult lifeform, and present that map in a clickable, navigable display — a sort of Google Earth for developmental biology.
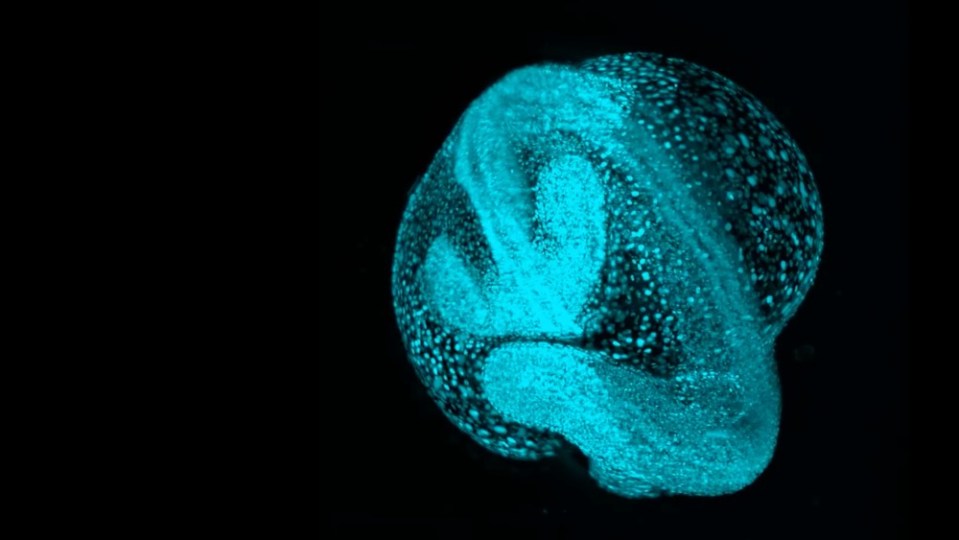
Now, in a new paper in Cell, researchers at the Chan Zuckerberg Biohub San Francisco (CZ Biohub SF) are unveiling the latest advancements in that effort. “Zebrahub” is a state-of-the-art cell atlas that combines high-resolution time-lapse videos of newly emerging cells in zebrafish embryos with extensive data on which genes are switched on and off as individual cells navigate to their eventual stations and “decide” what role they will ultimately play in the body of the adult fish.
A freshwater species native to South Asia, zebrafish as adults rarely exceed two inches in length, and they are a long-established model for developmental research relevant to human health. Around 70% of human genes have counterparts in zebrafish and, though we look quite different, as fellow vertebrates we share most of the same overall body plan in addition to the cellular and molecular processes by which various body parts initially form. Critically, zebrafish embryos are also mostly transparent and — unlike those of, say, mice — develop outside the mother, making it possible for scientists to observe their early growth in detail under a microscope.
Creating Zebrahub, which is free to all and includes built-in analytical tools designed for biologists, required building a suite of new instruments and software. It’s the most comprehensive atlas of its kind and, as the researchers write in the paper, an important step towards “ushering in a new era for developmental and evolutionary biology.”
“How a lifeform goes from a single cell to an entire body is one of biology’s biggest mysteries,” says senior author Loïc Royer, leader of the Organismal Architecture group and director of imaging AI at CZ Biohub SF. “With Zebrahub, we’ve created possibly the most detailed map of that process ever.”