By the end of this video, you should have a sense of how pathogen diversity accumulates during an infection, and how this diversity can be passed on, providing a signal of infection and transmission within a pathogen genome sequence.
Take-home messages
- Mutations can be deleterious, neutral, or beneficial.
- Within-host pathogen diversity is derived when pathogens make errors during replication, making many progeny with slightly different genome sequences.
- The frequency of all of these different lineages changes over the course of the individual’s infection. Some lineages might be purged from the population (e.g. when the mutation(s) they carry are lethal or deleterious), some lineages might remain at roughly the same frequency, and some lineages might rise in frequency, either through chance or because the mutation(s) they carry confer a benefit to the virus.
- At the time of a transmission event, the within-host diversity of the donor is sampled and transmitted to initiate the infection in the recipient.
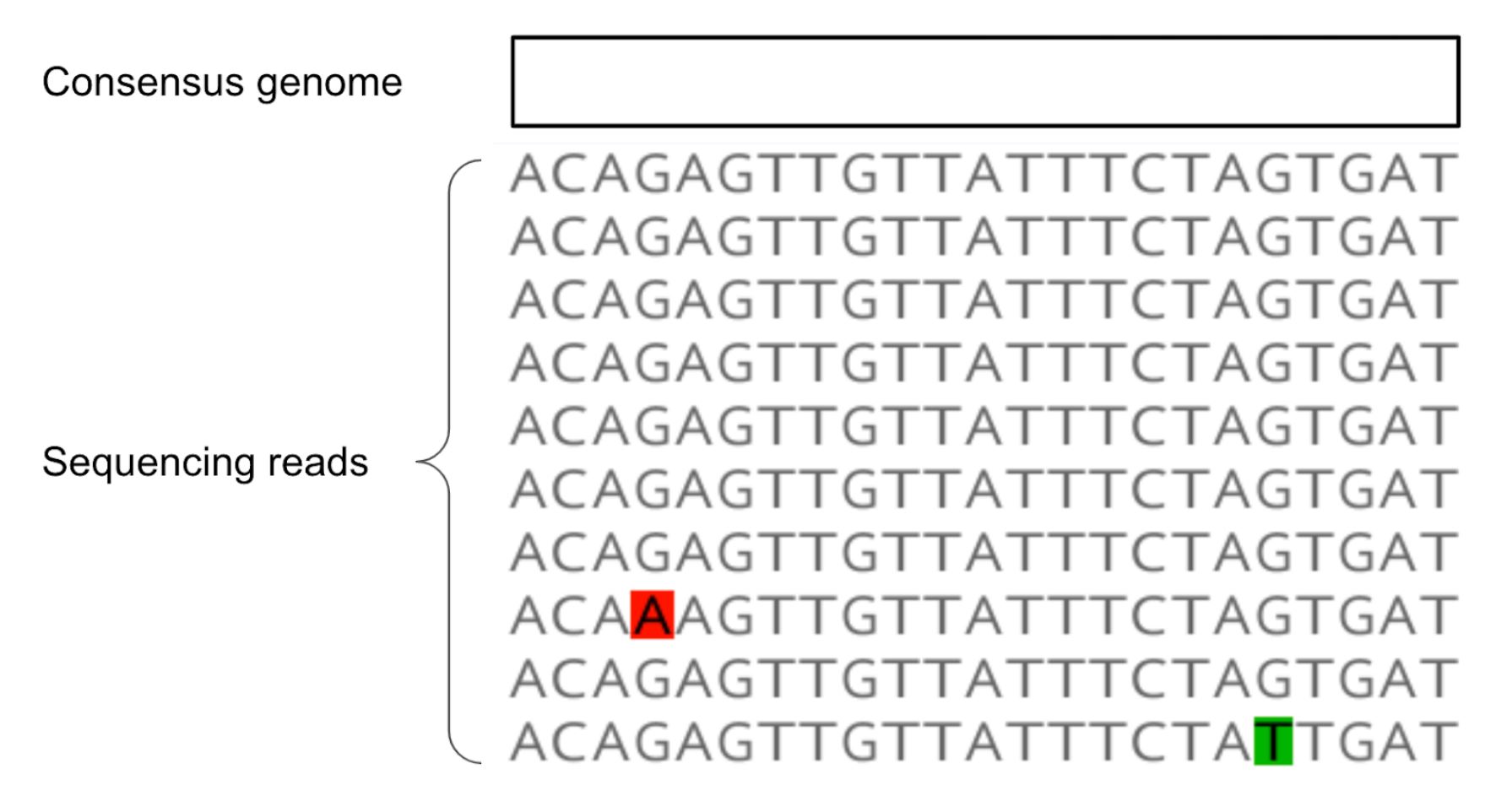
- When we sequence a clinical sample collected from an infected individual, we most commonly generate a consensus genome sequence. The consensus genome is a summary that describes which nucleotide was observed at the highest frequency in each site in the genome.
- Many of the mutations that have naturally occurred during the viral replication process will be present at such low frequency within an infection that they would be hard to distinguish from PCR errors or sequencing errors. This means that much of the within-host variation (changes to the genome sequence that occurs in nature) will not be observed in consensus genomes (your data).
- This means that even though mutations are always occurring over the course of an individual’s infection, we may not actually see a change in the consensus genome that summarizes two linked infections.
- The rate at which we observe substitutions varies depending on the pathogen’s mutation rate and also external forces such as the strength of selection or the density and frequency of sampling.
Questions
- We discussed that the consensus genome represents a summary of the most frequently observed nucleotides at each site in the sequence. Take a look at the figure below, which shows multiple sequencing reads mapped to a reference genome. Given the observed sequencing reads, what would the consensus genome sequence be?
- You observe two genome sequences that are genetically diverged from each other. Do you think these sequences were sampled from cases that are epidemiologically-linked? Why or why not?
- You observe two genome sequences that have identical sequences. Can you be sure that these viruses were sampled from a direct infection pair (one case directly infected the other case)?



