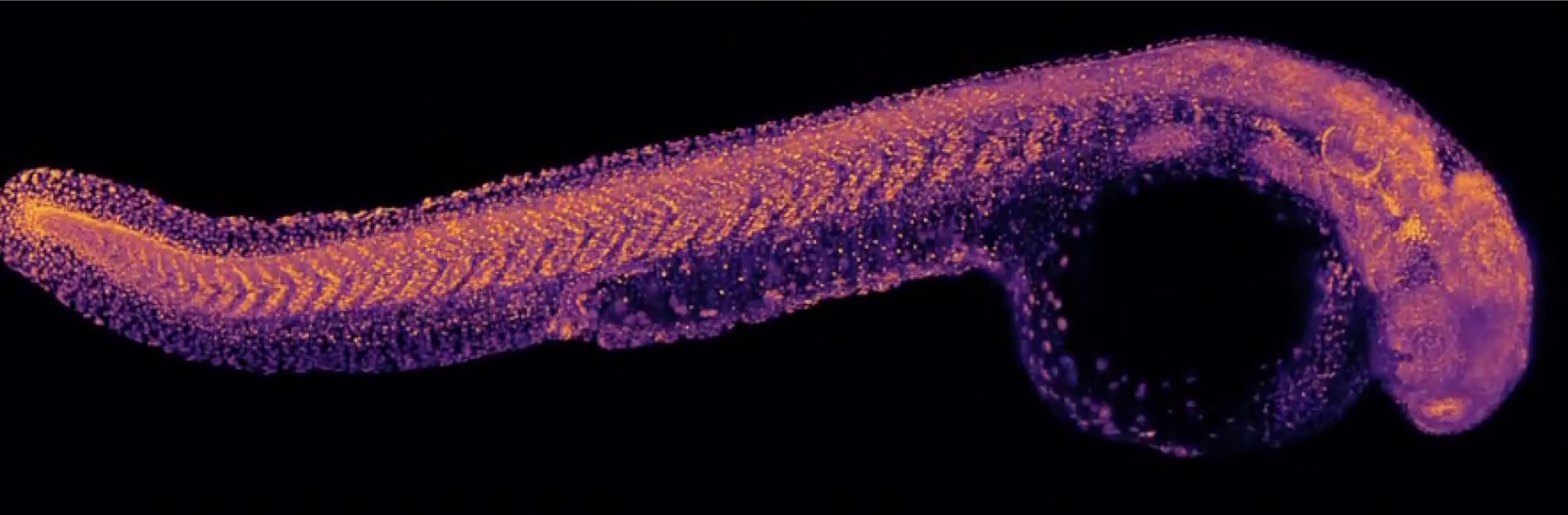
Everything everywhere all at once: ‘Zebrahub’ tracks development like never before
A ‘Google Earth’ of embryology, CZ Biohub San Francisco’s zebrafish cell atlas brings a new vision to developmental biology
READ MORE

With New Omega Tool, Scientists Can Rapidly Analyze Complex Biological Images through AI-Powered ‘Conversations’
In a new research article, scientists at CZ Biohub SF describe Omega, an open-source software tool that significantly advances the field of bioimage analysis. Omega harnesses the power of large language models (LLMs) to enable scientists to process and analyze biological images through natural language conversations rather than having to issue formal commands or write code.
READ MORE

The Tiny Zebrafish Hotel with Merlin Lange, Loïc Royer, and Shruthi VijayKumar
We enjoyed hosting podcaster Nate Butkus and his parents at our lab in San Francisco!
Listen to the podcast

Creator of Biohub’s ‘cytoself’ protein-localization method named 2022 STAT Wunderkind
Kobayashi cited as among ‘the brightest young minds’ in research.
Read more

Calling all LEGO builders! Put your skills to use assembling a light-sheet microscope
CZ Biohub’s game-changing microscope seeks to inspire an international community of builders.
Read more

What New Cell Biology Can AI Reveal Just by Looking at Images? A Lot!
As reported in Nature Methods, the algorithm, dubbed “cytoself,” provides rich, detailed information on protein location and function within a cell. This capability could quicken research time for cell biologists and eventually be used to accelerate the process of drug discovery and drug screening.
Read more

Python power-up: new image tool visualizes complex data
The image viewing and analysis software napari has filled a gap in the programming language’s scientific ecosystem.
Read More at Nature

Self-Supervised Deep-Learning Encodes High-Resolution Features of Protein Subcellular Localization
Elucidating the diversity and complexity of protein localization is essential to fully understand cellular architecture. Here, we present cytoself, a deep learning-based approach for fully self-supervised protein localization profiling and clustering.
Read More