The Tabula Projects
Reference transcriptomes at single-cell resolution
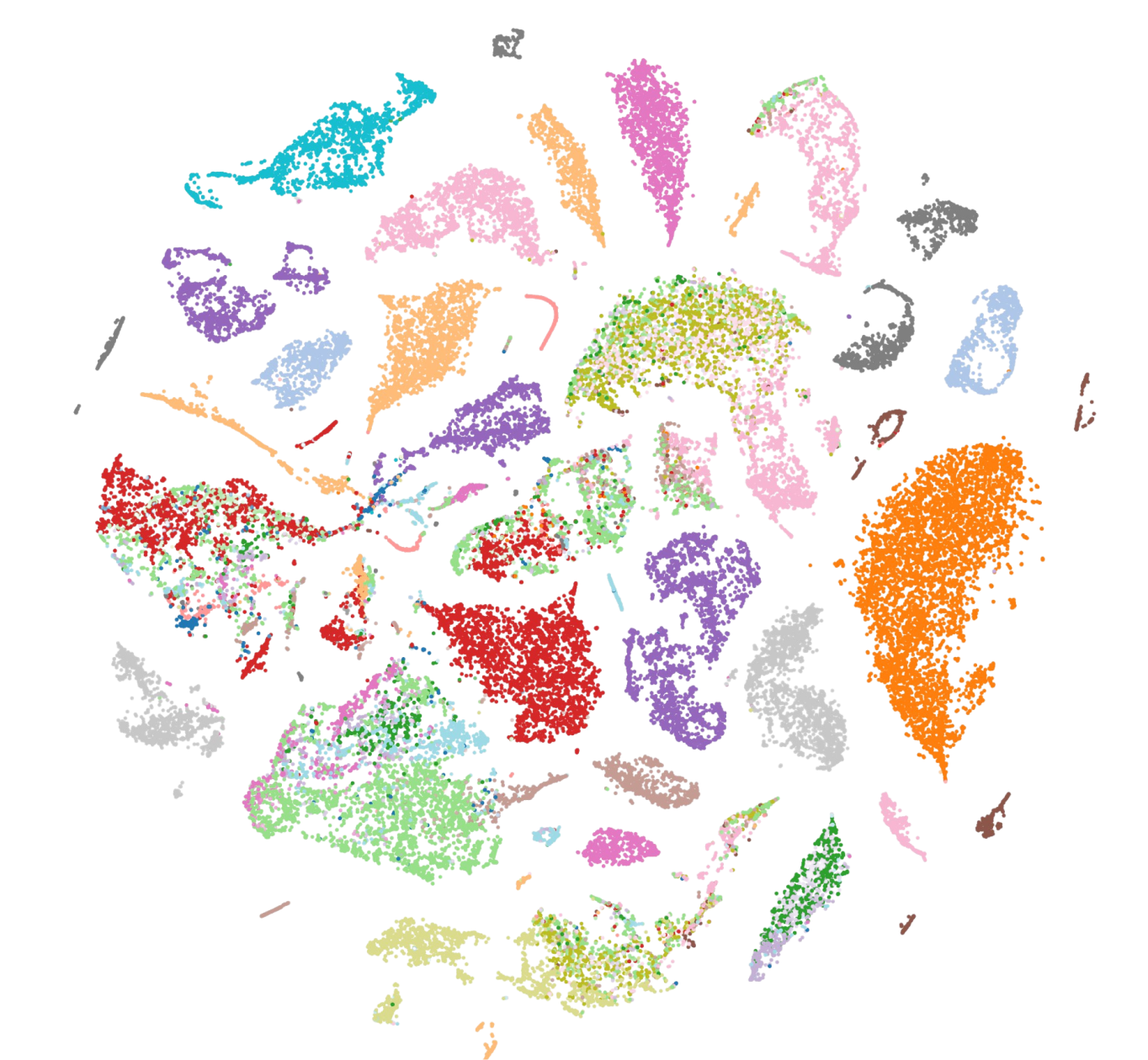
Reference transcriptomes at single-cell resolution
The CZ Biohub Tabula projects seek to explore these questions by mapping each cell type in whole organisms. The resulting cell atlases, or Tabulae, are essential companions to genomes, as they show which genes are expressed in a given cell and how this expression dictates the specialized functions of the many cell types that make up tissues and organs. They also help us understand what happens over the lifetime of an organism and when things go awry during disease.
These cell atlases are built using two state-of-the-art technologies: single-cell transcriptomics and the computational approaches necessary to analyze sequencing data from millions of reads in hundreds of thousands of cells. The Tabulae are assembled through a highly collaborative effort involving tissue experts, pathologists, cell and molecular biologists, and computational biologists.
While this work continues, CZ Biohub and its collaborators have produced the first whole-organism cell atlases for mice (Tabula Muris); aging mice (Tabula Muris Senis); the mouse lemur, a non-human primate (Tabula Microcebus); the fruit fly (Tabula Drosophilae); and humans (Tabula Sapiens). These rich data sets are providing new insight into evolution, cell physiology, and organismal biology in health and disease.
users of Tabula web portals
downloads of published mouse Tabula data sets
annotated cells in Tabula projects from human, mouse, fruit fly, and lemur
Tabula Sapiens is a single-cell transcriptomic atlas of nearly 500,000 cells from 24 different tissues and organs across 14 human donors. This resource provides a rich molecular characterization of more than 800 cell types, their distribution across tissues, and detailed information about tissue-specific variation in gene expression, representing the first draft of a broadly useful reference tool to understand human biology. Tabula Sapiens was funded by the Chan Zuckerberg Initiative as part of its single-cell biology program.

Zebrahub is a multimodal, single-cell RNA sequencing atlas of vertebrate development at single-embryo resolution, using zebrafish as a model organism. The first Zebrahub dataset, of approximately 120,000 cells, spans 10 developmental stages: from end-of-gastrulation embryos to 10-day larvae (bud-stage, 5-, 10-, 15-, 20-, 30-somites stages, as well as 2-, 3-, 5- and 10-days post-fertilization). Four embryos were sequenced per time point. We strive to achieve the highest possible quality; in that context, we expect the Zebrahub dataset to evolve to include more stages and higher data resolution. The Royer Group leads this work in collaboration with CZ Biohub’s Data Science and Genomics Platforms.

Tabula Muris is a compendium of single-cell transcriptome data from the mouse derived from nearly 100,000 cells from 20 organs and tissues. The data allow for direct and controlled comparison of gene expression in cell types shared between tissues, such as immune cells from distinct anatomical locations.

Tabula Muris Senis provides a rich resource to define the molecular signatures of the aging process in the mouse. It includes data from diverse cell types in 23 tissues and organs across the lifespan, and documents cell-specific changes that occur across multiple cell types and organs, as well as age-related changes in the cellular composition of different organs.

Tabula Microcebus comprises data from more than 220,000 single cells representing more than 750 defined cell types from 27 organs of the mouse lemur (Microcebus murinus). The atlas provides a cellular and molecular foundation for studying this primate model organism, and a general approach for establishing other emerging model organisms. The Tabula Microcebus is the result of an international collaborative effort involving the CZ Biohub, many groups at Stanford, the CNRS-MNHN (Musėum National d’Histoire Naturelle in Brunoy, France), and groups at Hong Kong University of Science and Technology, Cornell University, A*STAR (Agency for Science, Technology and Research in Singapore), and UC San Francisco.

The Tabula Drosophilae, or Fly Cell Atlas project, sequenced 580,000 cells, annotated by 42 international labs, and provides a molecular characterization of more than 250 cell types covering all tissues of the adult fruit fly, Drosophila melanogaster. The project was completed by a large international consortium driven by Hongjie Li of Baylor University and Liqun Luo of Stanford University, and supported by several key labs at KU Leuven, EPFL (Lausanne), Stanford University, Harvard University, the National Institutes of Health, and CZ Biohub, among other collaborators.
The COVID Tissue Atlas enables the investigation of both cell type-specific and cross-organ transcriptional responses to COVID-19, providing insights into the molecular networks affected by the disease and highlighting novel potential targets for therapies and drug development.
Subscribe to our bimonthly Nexus newsletter, and receive other updates from CZ Biohub Network.
Cookies and JavaScript are required to access this form.